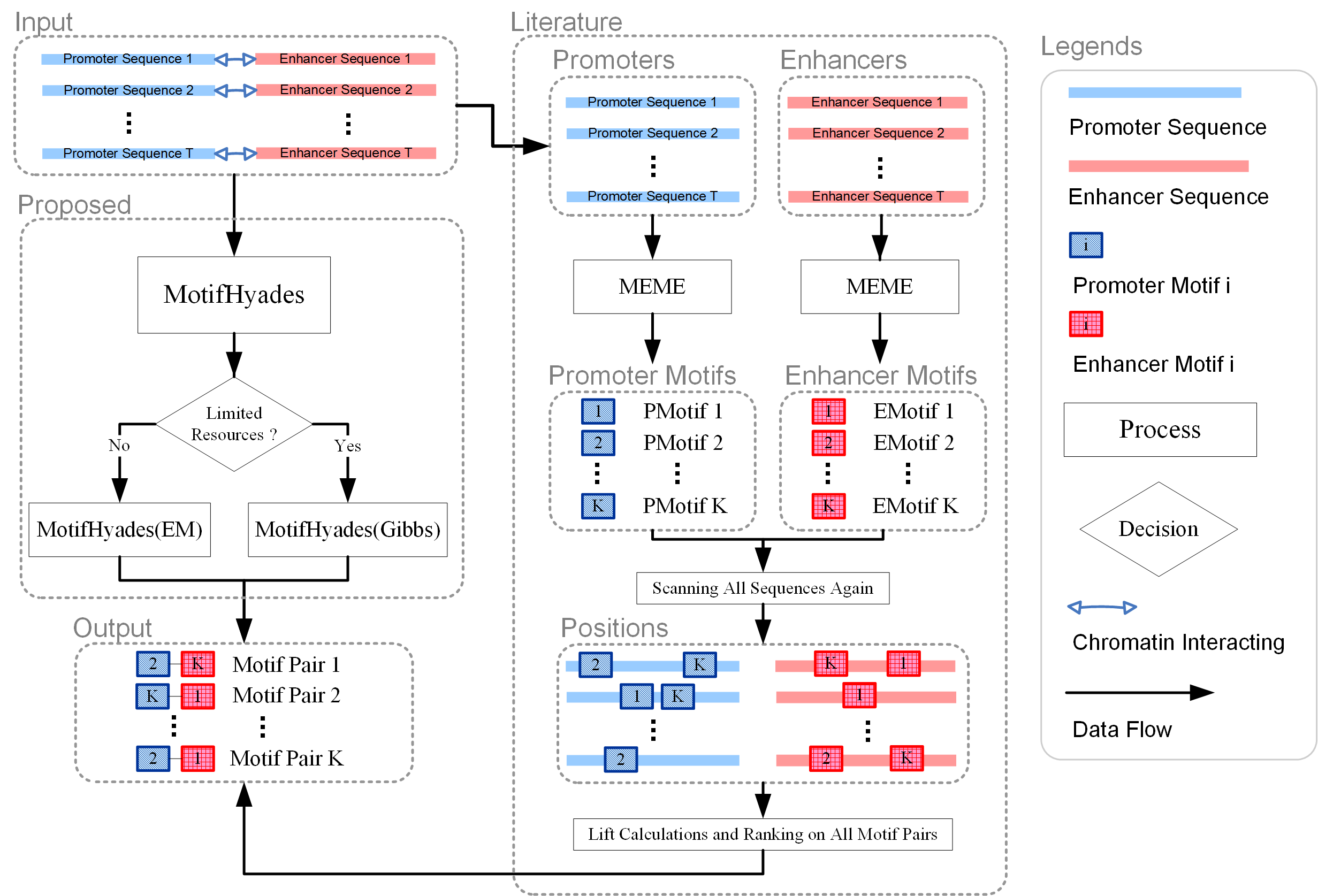
FAQ
What is MotifHyades ?
What is MCR ?
MCR stands for Matlab Compiler Runtime. If your machine does not have Matlab, you need to install MCR to execute MotifHyades. MCR can be downloaded from the internet easily. In particular, we advise you to download the same version indicated in the "Downloads" section.
Is there any demo ?
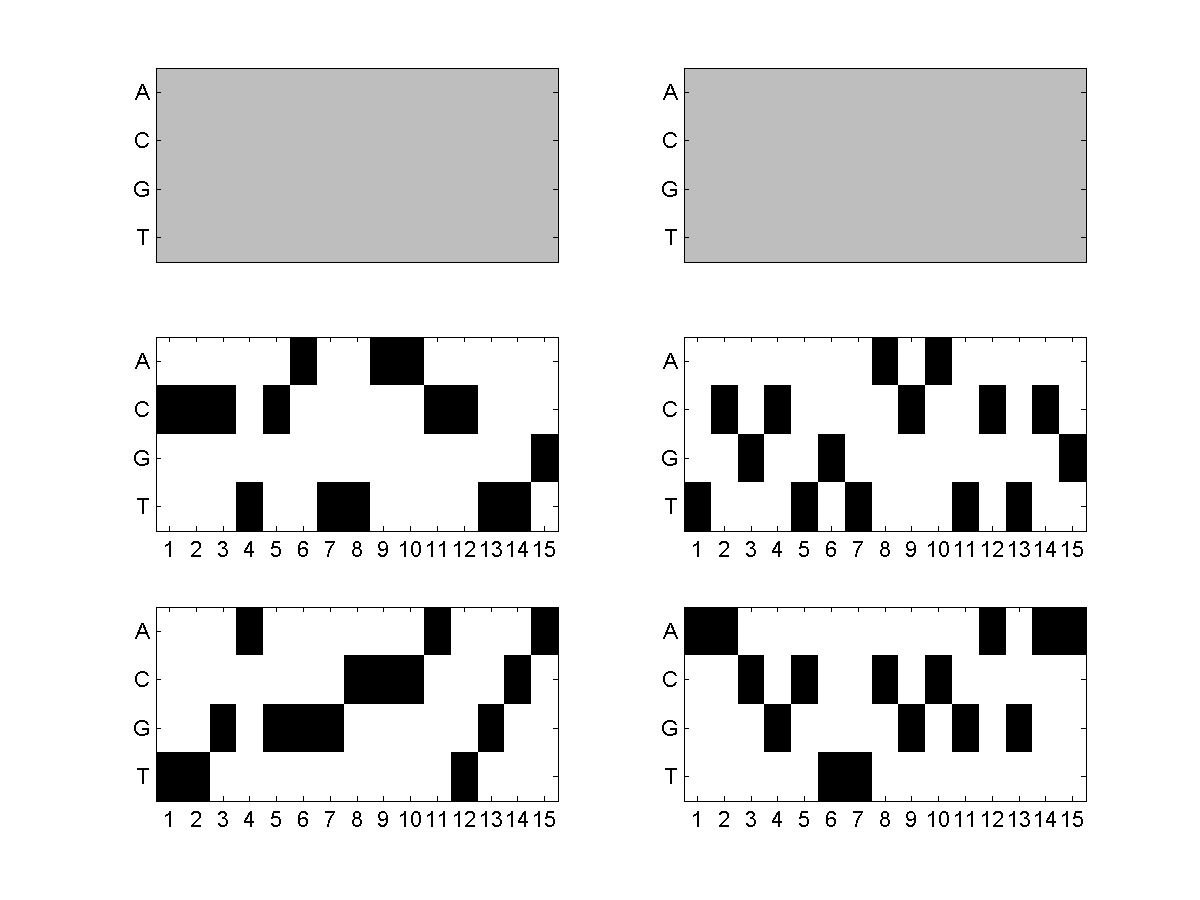
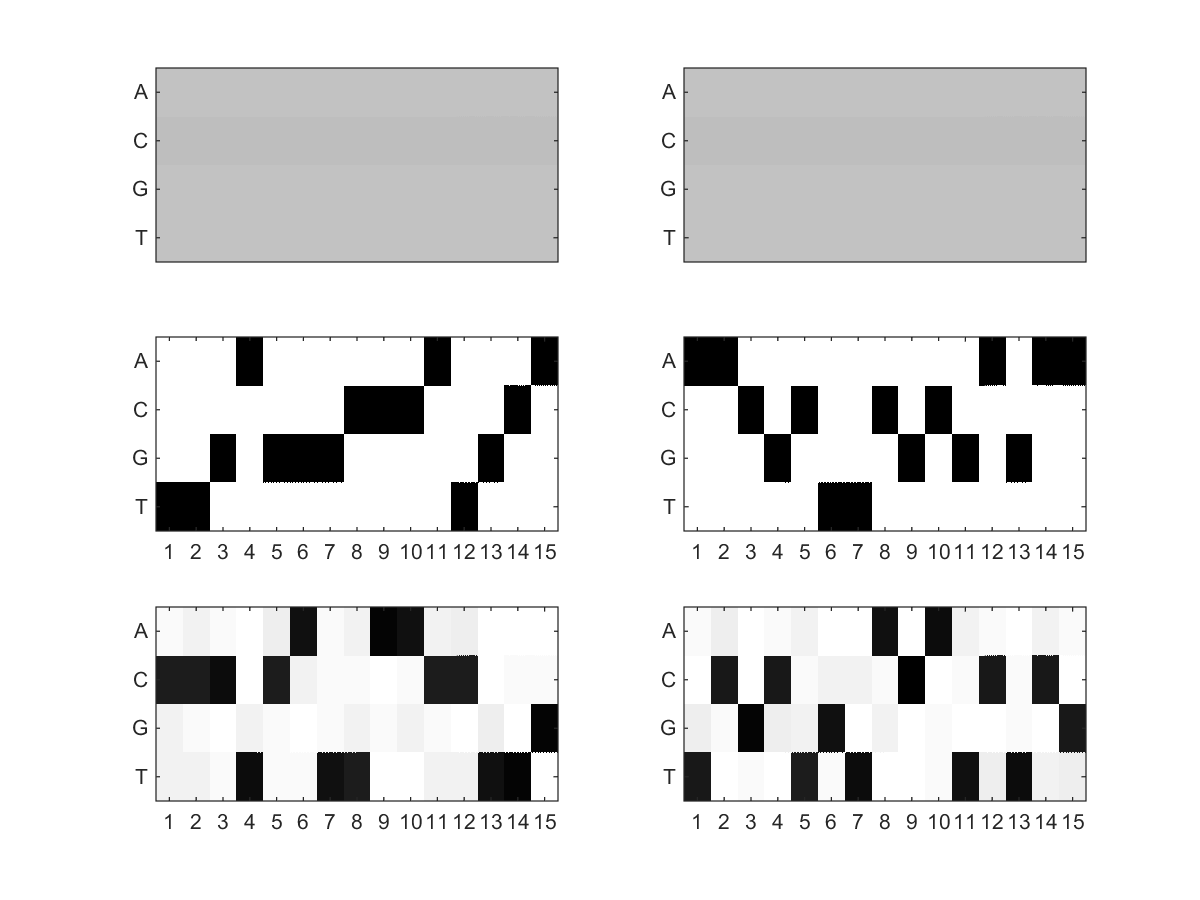
By default, a small testing dataset (SequenceSet1.fasta and SequenceSet2.fasta) is zipped with the MotifHyades executables in the "Downloads" section. Once downloaded, you can simply change your current directory to it and type "MotifHyades SequenceSet1.fasta SequenceSet2.fasta" to run a MotifHyades demo on the testing dataset (which has 2 DNA motif pairs to be discovered from the 100 sequence pairs). After the run, you will see the 2 DNA motif pairs discovered by MotifHyades. Details can be found in the result folders "MotifHyades_images" and "MotifHyades_results" generated; for instance, the following is the comparison between the actual 2 DNA motif pairs (left picture) and those discovered by MotifHyades (right picture). As you can see, the 2 DNA motif pairs discovered by MotifHyades (right picture) match well with the actual answer (left picture) except the discovery order which is normal in de novo motif pair discovery as we did not know the pair order in advance.
More data ?
Public genome annotation data can be accessed through ENCODE consortium and Gene Expression Omnibus (GEO).
